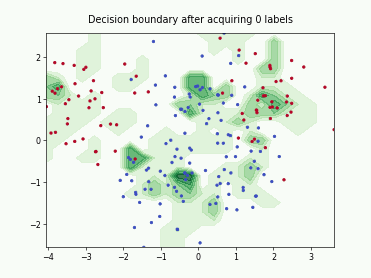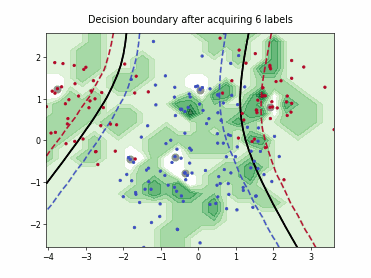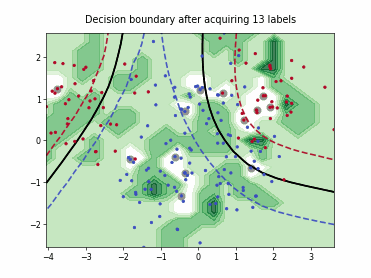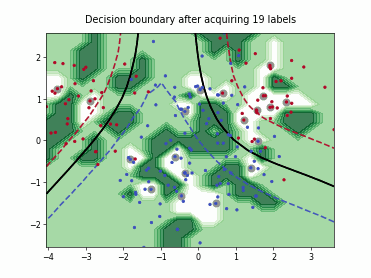
ProbCover#
- class skactiveml.pool.ProbCover(n_classes=None, deltas=None, alpha=0.95, cluster_algo=<class 'sklearn.cluster._kmeans.KMeans'>, cluster_algo_dict=None, n_cluster_param_name='n_clusters', distance_func=<function pairwise_distances>, missing_label=nan, random_state=None)[source]#
Bases:
SingleAnnotatorPoolQueryStrategyProbability Coverage (ProbCover)
This class implements the Probability Coverage (ProbCover) query strategy [1], which selects batch_size unlabeled points to maximize empirical coverage under a fixed radius delta in the embedding space, treating points within delta of any labeled sample as already covered and greedily adding the candidate samples that covers the most new samples at each step. It chooses delta via a purity criterion estimated from unlabeled data, prioritizes dense regions, and does not use predictive uncertainty.
- Parameters:
- n_classesNone or int, default=None
This parameter is used to determine the delta value. If n_classes=None, the number of classes is extracted from the given labels. If this extracted number of classes is below 2, n_classes=2 is used as a fallback.
- deltasNone or array-like of shape (n_deltas,), default=None
List of deltas (ball radii) to be tested for finding the maximum value satisfying a sample coverage >= alpha. If no value in deltas satisfies this constraint, a warning is raised where the minimum delta value is used. If deltas=None, the values np.arange(0.1, 2.1, 0.1) are used.
- alphafloat in (0, 1), alpha=0.95
Minimum coverage as a constraint for the delta selection.
- cluster_algoClusterMixin.__class__, default=sklearn.cluster.KMeans
The cluster algorithm to be used for determining the best delta value.
- cluster_algo_dictdict, default=None
The parameters passed to the clustering algorithm cluster_algo, excluding the parameter for the number of clusters.
- n_cluster_param_namestring, default=”n_clusters”
The name of the parameter for the number of clusters.
- distance_funccallable, default=sklearn.metrics.pairwise_distances
Takes as input X to compute the distances between each pair of samples. This function can also only return the precomputed distances of each pair in X for speedup.
- missing_labelscalar or string or np.nan or None, default=np.nan
Value to represent a missing label.
- random_stateNone or int or np.random.RandomState, default=None
The random state to use.
References
[1]O. Yehuda, A. Dekel, G. Hacohen, and D. Weinshall. Active Learning Through a Covering Lens. In Adv. Neural Inf. Process. Syst., 2022.
Methods
Get metadata routing of this object.
get_params([deep])Get parameters for this estimator.
query(X, y[, candidates, batch_size, ...])Determines for which candidate samples labels are to be queried.
set_params(**params)Set the parameters of this estimator.
- get_metadata_routing()#
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)#
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- query(X, y, candidates=None, batch_size=1, return_utilities=False, update=False)[source]#
Determines for which candidate samples labels are to be queried.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
Training data set, usually complete, i.e., including the labeled and unlabeled samples.
- yarray-like of shape (n_samples,)
Labels of the training data set (possibly including unlabeled ones indicated by self.missing_label).
- candidatesNone or array-like of shape (n_candidates), dtype=int or array-like of shape (n_candidates, n_features), default=None
If candidates is None, the unlabeled samples from (X,y) are considered as candidates.
If candidates is of shape (n_candidates,) and of type int, candidates is considered as the indices of the samples in (X,y).
- batch_sizeint, default=1
The number of samples to be selected in one AL cycle.
- return_utilitiesbool, default=False
If True, also return the utilities based on the query strategy.
- Returns:
- query_indicesnumpy.ndarray of shape (batch_size)
The query indices indicate for which candidate sample a label is to be queried, e.g., query_indices[0] indicates the first selected sample. The indexing refers to the samples in X.
- utilitiesnumpy.ndarray of shape (batch_size, n_samples) or numpy.ndarray of shape (batch_size, n_candidates)
The utilities of samples after each selected sample of the batch, e.g., utilities[0] indicates the utilities used for selecting the first sample (with index query_indices[0]) of the batch. Utilities for labeled samples will be set to np.nan. The indexing refers to the samples in X.
- set_params(**params)#
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.