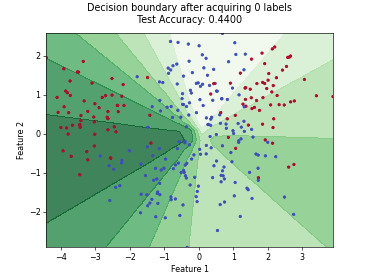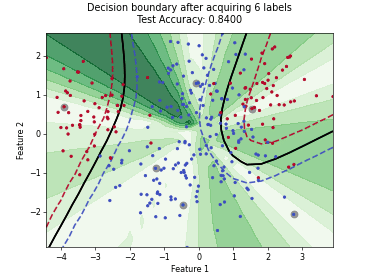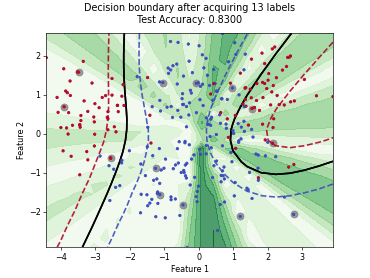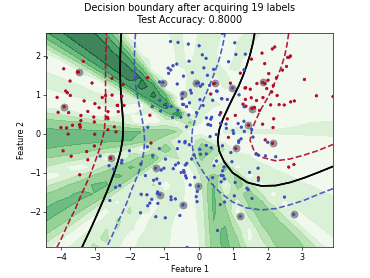
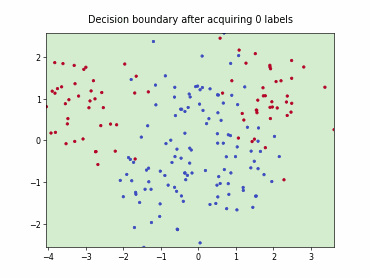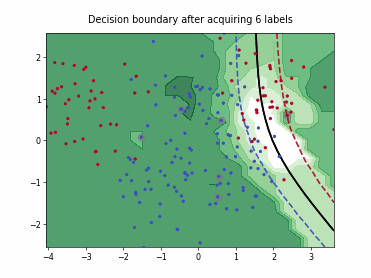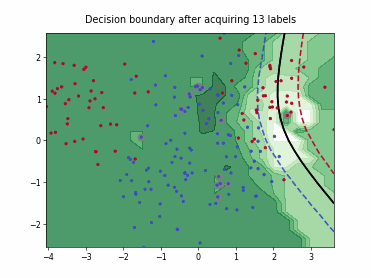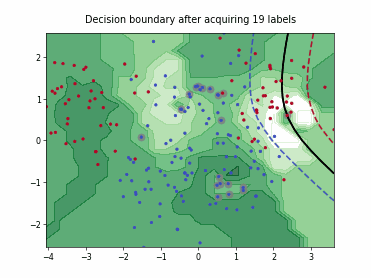
ParzenWindowClassifier#
- class skactiveml.classifier.ParzenWindowClassifier(n_neighbors=None, metric='rbf', metric_dict=None, classes=None, missing_label=nan, cost_matrix=None, class_prior=0.0, random_state=None)[source]#
Bases:
ClassFrequencyEstimatorParzen Window Classifier (PWC)
The “Parzen Window Classifier” (PWC) [1] is a simple and probabilistic classifier. This classifier is based on a non-parametric density estimation obtained by applying a kernel function.
- Parameters:
- classesarray-like of shape (n_classes), default=None
Holds the label for each class. If None, the classes are determined during the fit.
- missing_labelscalar or string or np.nan or None, default=np.nan
Value to represent a missing label.
- cost_matrixarray-like of shape (n_classes, n_classes), default=None
Cost matrix with cost_matrix[i,j] indicating cost of predicting class classes[j] for a sample of class classes[i]. Can be only set, if classes is not None.
- class_priorfloat or array-like of shape (n_classes,), default=0
Prior observations of the class frequency estimates. If class_prior is an array, the entry class_prior[i] indicates the non-negative prior number of samples belonging to class classes_[i]. If class_prior is a float, class_prior indicates the non-negative prior number of samples per class.
- metricstr or callable, default=’rbf’
The metric must be a valid kernel defined by the function sklearn.metrics.pairwise.pairwise_kernels.
- n_neighborsint or None, default=None
Number of nearest neighbours. Default is None, which means all available samples are considered.
- metric_dictdict, default=None
Any further parameters are passed directly to the kernel function. For the kernel ‘rbf’ we allow the use of mean bandwidth criterion [2] and use it when gamma is set to ‘mean’ (i.e., {‘gamma’: ‘mean’})..
- random_stateint or RandomState instance or None, default=None
Determines random number for predict method. Pass an int for reproducible results across multiple method calls.
- Attributes:
- classes_numpy.ndarray of shape (n_classes,)
Holds the label for each class after fitting.
- class_priornp.ndarray of shape (n_classes,)
Prior observations of the class frequency estimates. The entry class_prior_[i] indicates the non-negative prior number of samples belonging to class classes_[i].
- cost_matrix_np.ndarray of shape (classes, classes)
Cost matrix with cost_matrix_[i,j] indicating cost of predicting class classes_[j] for a sample of class classes_[i].
- X_np.ndarray of shape (n_samples, n_features)
The sample matrix X is the feature matrix representing the samples.
- V_np.ndarray of shape (n_samples, classes)
The class labels are represented by counting vectors. An entry V[i,j] indicates how many class labels of classes[j] were provided for training sample X_[i].
References
[1]O. Chapelle, “Active Learning for Parzen Window Classifier”, Proceedings of the Tenth International Workshop Artificial Intelligence and Statistics, 2005.
[2]Chaudhuri, A., Kakde, D., Sadek, C., Gonzalez, L., & Kong, S., “The Mean and Median Criteria for Kernel Bandwidth Selection for Support Vector Data Description” IEEE International Conference on Data Mining Workshops (ICDMW), 2017.
Methods
fit(X, y[, sample_weight])Fit the model using X as samples and y as class labels.
Get metadata routing of this object.
get_params([deep])Get parameters for this estimator.
predict(X, **kwargs)Return class label predictions for the test samples X.
predict_freq(X)Return class frequency estimates for the input samples X.
predict_proba(X, **kwargs)Return probability estimates for the test data X.
sample_proba(X[, n_samples, random_state])Samples probability vectors from Dirichlet distributions whose parameters alphas are defined as the sum of the frequency estimates returned by predict_freq and the class_prior.
score(X, y[, sample_weight])Return the mean accuracy on the given test data and labels.
set_fit_request(*[, sample_weight])Configure whether metadata should be requested to be passed to the
fitmethod.set_params(**params)Set the parameters of this estimator.
set_score_request(*[, sample_weight])Configure whether metadata should be requested to be passed to the
scoremethod.Attributes
- METRICS = ['additive_chi2', 'chi2', 'cosine', 'linear', 'poly', 'polynomial', 'rbf', 'laplacian', 'sigmoid', 'precomputed']#
- fit(X, y, sample_weight=None)[source]#
Fit the model using X as samples and y as class labels.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
The feature matrix representing the samples.
- yarray-like of shape (n_samples,)
It contains the class labels of the training samples.
- sample_weightarray-like of shape (n_samples), default=None
It contains the weights of the training samples’ class labels. It must have the same shape as y.
- Returns:
- selfParzenWindowClassifier,
The ParzenWindowClassifier is fitted on the training data.
- get_metadata_routing()#
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)#
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- predict(X, **kwargs)#
Return class label predictions for the test samples X.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
Input samples.
- Returns:
- ynumpy.ndarray of shape (n_samples,)
Predicted class labels of the test samples X.
- predict_freq(X)[source]#
Return class frequency estimates for the input samples X.
- Parameters:
- Xarray-like or shape (n_samples, n_features) or shape (n_samples, m_samples) if metric == ‘precomputed’
Input samples.
- Returns:
- Fnp.ndarray of shape (n_samples, classes)
The class frequency estimates of the input samples. Classes are ordered according to the attribute classes_.
- predict_proba(X, **kwargs)#
Return probability estimates for the test data X.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
Input samples.
- Returns:
- Parray-like of shape (n_samples, classes)
The class probabilities of the test samples. Classes are ordered according to self.classes_.
- sample_proba(X, n_samples=10, random_state=None)#
Samples probability vectors from Dirichlet distributions whose parameters alphas are defined as the sum of the frequency estimates returned by predict_freq and the class_prior.
- Parameters:
- Xarray-like of shape (n_test_samples, n_features)
Test samples for which n_samples probability vectors are to be sampled.
- n_samplesint, default=10
Number of probability vectors to sample for each X[i].
- random_stateint or numpy.random.RandomState or None, default=None
Ensure reproducibility when sampling probability vectors from the Dirichlet distributions.
- Returns:
- Parray-like of shape (n_samples, n_test_samples, n_classes)
There are n_samples class probability vectors for each test sample in X. Classes are ordered according to self.classes_.
- score(X, y, sample_weight=None)#
Return the mean accuracy on the given test data and labels.
- Parameters:
- Xarray-like of shape (n_samples, n_features)
Test samples.
- yarray-like of shape (n_samples,)
True labels for X.
- sample_weightarray-like of shape (n_samples,), default=None
Sample weights.
- Returns:
- scorefloat
Mean accuracy of self.predict(X) regarding y.
- set_fit_request(*, sample_weight: bool | None | str = '$UNCHANGED$') ParzenWindowClassifier#
Configure whether metadata should be requested to be passed to the
fitmethod.Note that this method is only relevant when this estimator is used as a sub-estimator within a meta-estimator and metadata routing is enabled with
enable_metadata_routing=True(seesklearn.set_config()). Please check the User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed tofitif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it tofit.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.Added in version 1.3.
- Parameters:
- sample_weightstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_weightparameter infit.
- Returns:
- selfobject
The updated object.
- set_params(**params)#
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
- set_score_request(*, sample_weight: bool | None | str = '$UNCHANGED$') ParzenWindowClassifier#
Configure whether metadata should be requested to be passed to the
scoremethod.Note that this method is only relevant when this estimator is used as a sub-estimator within a meta-estimator and metadata routing is enabled with
enable_metadata_routing=True(seesklearn.set_config()). Please check the User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed toscoreif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it toscore.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.Added in version 1.3.
- Parameters:
- sample_weightstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_weightparameter inscore.
- Returns:
- selfobject
The updated object.
Examples using skactiveml.classifier.ParzenWindowClassifier#
Batch Active Learning by Diverse Gradient Embedding (BADGE)
Fast Active Learning by Contrastive UNcertainty (FALCUN)
Monte-Carlo Expected Error Reduction (EER) with Log-Loss
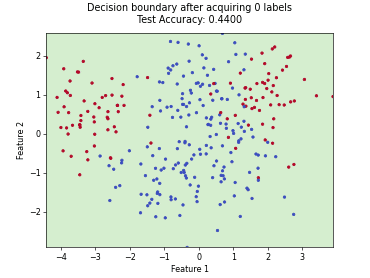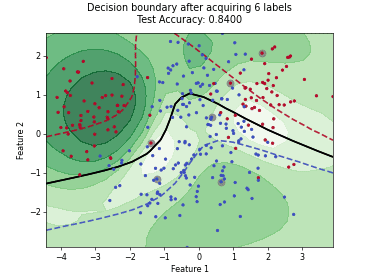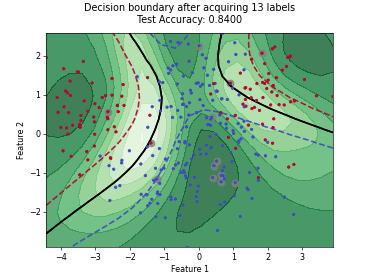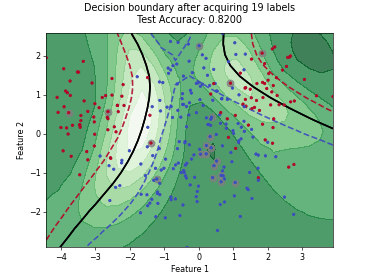
Monte-Carlo Expected Error Reduction (EER) with Misclassification-Loss
Query-by-Committee (QBC) with Kullback-Leibler Divergence
Querying Informative and Representative Examples (QUIRE)

Uncertainty Sampling with Expected Average Precision (USAP)
Cognitive Dual-Query Strategy with Fixed-Uncertainty
Cognitive Dual-Query Strategy with Random Sampling
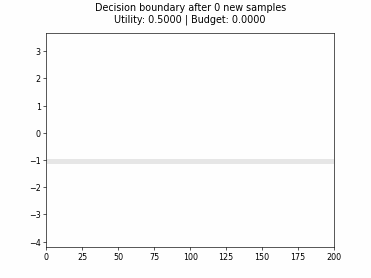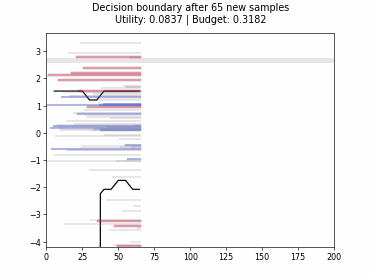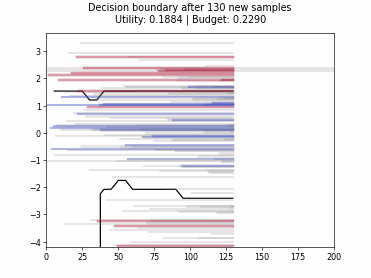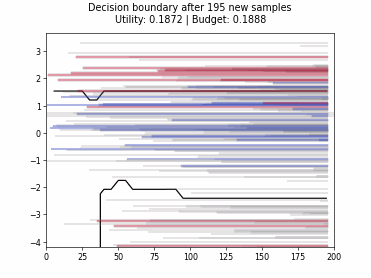
Cognitive Dual-Query Strategy with Randomized-Variable-Uncertainty
Cognitive Dual-Query Strategy with Variable-Uncertainty